Complexity: A Guided Tour - Melanie Mitchell (2009)
Part III. Computation Writ Large
Chapter 12. Information Processing in Living Systems
EVER SINCE SZILARD’S INSIGHT THAT information might be the savior of the second law of thermodynamics from the attack of Maxwell’s demon, information and its cousin computation have increasingly infiltrated science. In many people’s minds information has taken on an ontological status equal to that of mass and energy—namely, as a third primitive component of reality. In biology in particular, the description of living systems as information processing networks has become commonplace. In fact, the term information processing is so widely used that one would think it has a well-understood, agreed-upon meaning, perhaps built on Shannon’s formal definition of information. However, like several other central terms of complex systems science, the concept of information processing tends to be ill-defined; it’s often hard to glean what is meant by information processing or computation when they are taken out of the precise formal context defined by Turing machines and von Neumann-style computers. The work described in the previous chapter was an attempt to address this issue in the context of cellular automata.
The purpose of this chapter is to explore the notion of information processing or computation in living systems. I describe three different natural systems in which information processing seems to play a leading role—the immune system, ant colonies, and cellular metabolism—and attempt to illuminate the role of information and computation in each. At the end I attempt to articulate some common qualitative principles of information processing in these and other decentralized systems.
What Is Information Processing?
Let me quote myself from chapter 10: “In what sense do natural systems ‘compute’? At a very general level, one might say that computation is what a complex system does with information in order to succeed or adapt in its environment. But can we make this statement more precise? Where is the information, and what exactly does the complex system do with it?” These questions may seem straightforward, but exploring them will quickly force us to dip our feet into some of the murkiest waters in complex systems science.
When we say a system is processing information or computing (terms which, for now, I will use synonymously), we need to answer the following:
· What plays the role of “information” in this system?
· How is it communicated and processed?
· How does this information acquire meaning? And to whom? (Some will disagree with me that computation requires meaning of some sort, but I will go out on a limb and claim it does.)
INFORMATION PROCESSING IN TRADITIONAL COMPUTERS
As we saw in chapter 4, the notion of computation was formalized in the 1930s by Alan Turing as the steps carried out by a Turing machine on a particular input. Ever since then, Turing’s formulation has been the basis for designing traditional von-Neumann-style programmable computers. For these computers, questions about information have straightforward answers. We can say that the role of information is played by the tape symbols and possible states of the tape head. Information is communicated and processed by the tape head’s actions of reading from and writing to the tape, and changing state. This is all done by following the rules, which constitute the program.
We can view all programs written for traditional computers at (at least) two levels of description: a machine-code level and a programming level. The machine-code level is the set of specific, step-by-step low-level instructions for the machine (e.g., “move the contents of memory location n to CPU register j,” “perform an or logic operation on the contents of CPU registers j and i and store the result in memory location m,” and so on. The programming level is the set of instructions in a high-level language, such as BASIC or Java, that is more understandable to humans (e.g., “multiply the value of the variable half_of_total by 2 and store the result in the variable total,” etc.). Typically a single high-level instruction corresponds to several low-level instructions, which may be different on different computers. Thus a given high-level program can be implemented in different ways in machine code; it is a more abstract description of information processing.
The meaning of the input and output information in a Turing machine comes from its interpretation by humans (programmers and users). The meaning of the information created in intermediate steps in the computation also comes from its interpretation (or design) by humans, who understand the steps in terms of commands in a high-level programming language. This higher level of description allows us to understand computations in a human-friendly way that is abstracted from particular details of machine code and hardware.
INFORMATION PROCESSING IN CELLULAR AUTOMATA
For non-von-Neumann-style computers such as cellular automata, the answers are not as straightforward. Consider, for example, the cellular automaton described in the previous chapter that was evolved by a genetic algorithm to perform majority classification. Drawing an analogy with traditional computers, we could say that information in this cellular automaton is located in the configurations of states in the lattice at each time step. The input is the initial configuration, the output is the final configuration, and at each intermediate time step information is communicated and processed within each neighborhood of cells via the actions of the cellular automaton rule. Meaning comes from the human knowledge of the task being performed and interpretation of the mapping from the input to the output (e.g., “the final lattice is all white; that means that the initial configuration had a majority of white cells”).
Describing information processing at this level, analogous to “machine code,” does not give us a human-friendly understanding of how the computation is accomplished. As in the case of von-Neumann-style computation, we need a higher-level language to make sense of the intermediate steps in the computation and to abstract from particular details of the underlying cellular automaton.
In the previous chapter I proposed that particles and their interactions are one approach toward such a high-level language for describing how information processing is done in cellular automata. Information is communicated via the movement of particles, and information is processed via collisions between particles. In this way, the intermediate steps of information processing acquire “meaning” via the human interpretation of the actions of the particles.
One thing that makes von-Neumann-style computation easier to describe is that there is a clear, unambiguous way to translate from the programming level to the machine code level and vice versa, precisely because these computers were designed to allow such easy translation. Computer science has given us automatic compilers and decompilers that do the translation, allowing us to understand how a particular program is processing information.
For cellular automata, no such compilers or decompilers exist, at least not yet, and there is still no practical and general way to design high-level “programs.” Relatively new ideas such as particles as high-level information-processing structures in cellular automata are still far from constituting a theoretical framework for computation in such systems.
The difficulties for understanding information processing in cellular automata arise in spades when we try to understand information processing in actual living systems. My original question, “In what sense do natural systems ‘compute’?” is still largely unanswered, and remains a subject of confusion and thorny debate among scientists, engineers, and philosophers. However, it is a tremendously important question for complex systems science, because a high-level description of information processing in living systems would allow us not only to understand in new and more comprehensive ways the mechanisms by which particular systems operate, but also to abstract general principles that transcend the overwhelming details of individual systems. In essence, such a description would provide a “high-level language” for biology.
The rest of this chapter tries to make sense of these ideas by looking at real examples.
The Immune System
I gave a quick description of the immune system way back in chapter 1. Now let’s look a bit more in depth at how it processes information in order to protect the body from pathogens—viruses, bacteria, parasites, and other unwelcome intruders.
To recap my quick description, the immune system consists of trillions of different cells and molecules circulating in the body, communicating with one another through various kinds of signals.
Of the many many different types of cells in the immune system, the one I focus on here is the lymphocyte (a type of white blood cell; see figure 12.1). Lymphocytes are created in the bone marrow. Two of the most important types of lymphocytes are B cells, which release antibodies to fight viruses and bacteria, and T cells, which both kill invaders and regulate the response of other cells.
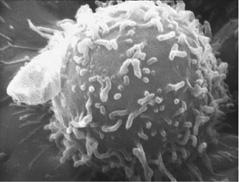
FIGURE 12.1. A human lymphocyte, whose surface is covered with receptors that can bind to certain shapes of molecules that the cell might encounter. (Photograph from National Cancer Institute [http://visualsonline.cancer.gov/details.cfm? imageid=1944].)
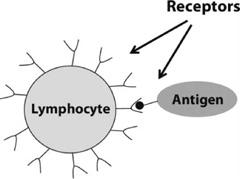
FIGURE 12.2. An illustration of a lymphocyte (here a B cell) receptor binding to an antigen.
Each cell in the body has molecules on its surface called receptors. As the name implies, these molecules are a means by which cells receive information. The information is in the form of other molecules that chemically bind to the receptor molecules. Whether or not a receptor binds to a particular molecule depends on whether their physical shapes match sufficiently.
A lymphocyte’s surface is covered with identical receptors that bind to a particular range of molecular shapes. If a lymphocyte happens by chance to encounter a special pathogen molecule (called an “antigen”) whose shape fits the lymphocyte’s receptors, then the molecule binds to one of the lymphocyte’s receptors, and the lymphocyte is said to have “recognized” that antigen—the first step in killing off pathogens. The binding can be strong or weak, depending on how closely the molecule’s shape fits the receptor. This process is illustrated in figure 12.2.
The main problem facing the immune system is that it doesn’t know ahead of time what pathogens will invade the body, so it can’t “predesign” a set of lymphocytes with receptors that will bind strongly to just the right shapes. What’s more, there are an astronomical number of possible pathogens, so the immune system will never be able to generate enough lymphocytes at any one time to take care of every eventuality. Even with all the many millions of different lymphocytes the body generates per day, the world of pathogens that the system will be likely to encounter is much bigger.
Here’s how the immune system solves this problem. In order to “cover” the huge space of possible pathogen shapes in a reasonable way, the population of lymphocytes in the body at any given time is enormously diverse. The immune system employs randomness to allow each individual lymphocyte to recognize a range of shapes that differs from the range recognized by other lymphocytes in the population.
In particular, when a lymphocyte is born, a novel set of identical receptors is created via a complicated random shuffling process in the lymphocyte’s DNA. Because of continual turnover of the lymphocyte population (about ten million new lymphocytes are born each day), the body is continually introducing lymphocytes with novel receptor shapes. For any pathogen that enters the body, it will just be a short time before the body produces a lymphocyte that binds to that pathogen’s particular marker molecules (i.e., antigens), though the binding might be fairly weak.
Once such a binding event takes place, the immune system has to figure out whether it is indicative of a real threat or is just a nonthreatening situation that can be ignored. Pathogens are harmful, of course, because once they enter the body they start to make copies of themselves in large numbers. However, launching an immune system attack can cause inflammation and other harm to the body, and too strong an attack can be lethal. The immune system as a whole has to determine whether the threat is real and severe enough to warrant the risk of an immune response harming the body. The immune system will go into high-gear attack mode only if it starts picking up a lot of sufficiently strong binding events.
The two types of lymphocytes, B and T cells, work together to determine whether an attack is warranted. If the number of strongly bound receptors on a B cell exceeds some threshold, and in the same time frame the B cell gets “go-ahead” signals from T cells with similarly bound receptors, the B cell is activated, meaning that it now perceives a threat to the body (figure 12.3). Once activated, the B cell releases antibody molecules into the bloodstream. These antibodies bind to antigens, neutralize them, and mark them for destruction by other immune system cells.
The activated B cell then migrates to a lymph node, where it divides rapidly, producing large numbers of daughter cells, many with mutations that alter the copies’ receptor shapes. These copies are then tested on antigens that are captured in the lymph node. The cells that do not bind die after a short time.
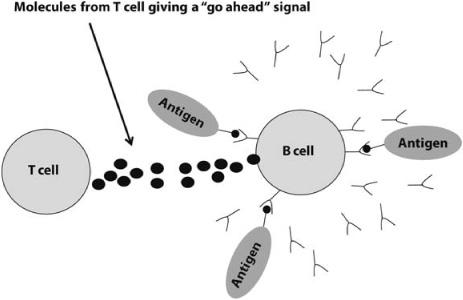
FIGURE 12.3. Illustration of activation of a B cell via binding and “go-ahead” signal from a T cell. This signal prompts the B cell to produce and release antibodies (y-shaped molecules).
The surviving copies are unleashed into the bloodstream, where some of them encounter and bind to antigens, in some cases more strongly than did their mother B cell. These activated daughter B cells also migrate to lymph nodes and create mutated daughters of their own. This massive production of lymphocytes is one reason that your lymph nodes often feel swollen when you are sick.
This cycle continues, with the new B cells that best match antigens themselves producing the most daughter cells. In short, this is a process of natural selection, in which collections of B cells evolve to have receptor shapes that will bind strongly to a target antigen. This results in a growing arsenal of antibodies that have been “designed” via selection to attack this specific antigen. This process of detection and destruction typically takes from a few days to weeks to eradicate the corresponding pathogen from the body.
There are at least two potential problems with this strategy. First, how does the immune system prevent lymphocytes from mistakenly attacking the body’s own molecules? Second, how does the immune system stop or tone down its attack if the body is being harmed too much as a result?
Immunologists don’t yet have complete answers to these questions, and each is currently an area of active research. It is thought that one major mechanism for avoiding attacking one’s own body is a process called negative selection. When lymphocytes are born they are not immediately released into the bloodstream. Instead they are tested in the bone marrow and thymus by being exposed to molecules of one’s own body. Lymphocytes that bind strongly to “self” molecules tend to be killed off or undergo “editing” in the genes that give rise to receptors. The idea is that the immune system should only use lymphocytes that will not attack the body. This mechanism often fails, sometimes producing autoimmune disorders such as diabetes or rheumatoid arthritis.
A second major mechanism for avoiding autoimmune attacks seems to be the actions of a special subpopulation of T cells called regulatory T cells. It’s not yet known exactly how these regulatory T cells work, but they do secrete chemicals that suppress the actions of other T cells. A third mechanism has been hypothesized to be the competition among B cells for a limited resource—a particular chemical named BAFF needed for B cells to survive. B cells that slip through the negative selection process and still bind strongly to self-molecules find themselves, due to their continual binding to self-molecules, in need of higher amounts of BAFF than non-self-binding B cells. Competition for this limited resource leads to the increased probability of death for self-binding B cells.
Even if the immune system is attacking foreign pathogens, it needs to balance the virulence of its attack with the obligation to prevent harm to the body as much as possible. The immune system employs a number of (mostly little understood) mechanisms for achieving this balance. Many of these mechanisms rely on a set of signaling molecules called cytokines. Harm to the body can result in the secretion of cytokines, which suppress active lymphocytes. Presumably the more harm being done, the higher the concentration of suppression cytokines, which makes it more likely that active cells will encounter them and turn off, thus regulating the immune system without suppressing it altogether.
Ant Colonies
As I described in chapter 1, analogies often have been made between ant colonies and the brain. Both can be thought of as networks of relatively simple elements (neurons, ants) from which emerge larger-scale information-processing behaviors. Two examples of such behavior in ant colonies are the ability to optimally and adaptively forage for food, and the ability to adaptively allocate ants to different tasks as needed by the colony. Both types of behavior are accomplished with no central control, via mechanisms that are surprisingly similar to those described above for the immune system.
In many ant species, foraging for food works roughly as follows. Foraging ants in a colony set out moving randomly in different directions. When an ant encounters a food source, it returns to the nest, leaving a trail made up of a type of signaling chemicals called pheromones. When other ants encounter a pheromone trail, they are likely to follow it. The greater the concentration of pheromone, the more likely an ant will be to follow the trail. If an ant encounters the food source, it returns to the nest, reinforcing the trail. In the absence of reinforcement, a pheromone trail will evaporate. In this way, ants collectively build up and communicate information about the locations and quality of different food sources, and this information adapts to changes in these environmental conditions. At any given time, the existing trails and their strengths form a good model of the food environment discovered collectively by the foragers (figure 12.4).
Task allocation is another way in which an ant colony regulates its own behavior in a decentralized way. The ecologist Deborah Gordon has studied task allocation in colonies of Red Harvester ants. Workers in these colonies divide themselves among four types of tasks: foraging, nest-maintenance, patrolling, and refuse-sorting work. The number of workers pursuing each type of task adapts to changes in the environment. Gordon found, for example, that if the nest is disturbed in some small way, the number of nest maintenance workers will increase. Likewise, if the food supply in the neighborhood is large and high quality, the number of foragers will increase. How does an individual ant decide which task to adopt in response to nestwide environmental conditions, even though no ant directs the decision of any other ant and each ant interacts only with a small number of other ants?

FIGURE 12.4. An ant trail. (Photograph copyright © by Flagstaffotos. Reproduced by permission.)
The answer seems to be that ants decide to switch tasks both as a function of what they encounter in the environment and as a function of the rate at which they encounter other ants performing different tasks. For example, an inactive ant—one not currently performing a task—that encounters a foreign object near the nest has increased probability of taking up nest-maintenance work. In addition, an inactive ant that encounters a high rate of nest-maintenance workers entering and leaving the nest will also have an increased probability of adopting the nest-maintenance task; the increased activity in some way signals that there are important nest maintenance tasks to be done. In a similar way, a nest-maintenance worker who encounters a high rate of foragers returning to the nest carrying seeds will have an increased probability of switching to foraging; the increased seed delivery signals in some way that a high-quality food source has been found and needs to be exploited. Ants are apparently able to sense, through direct contact of their antennae with other ants, what task the other ants have been engaged in, by perceiving specific chemical residues associated with each task.
Similar types of mechanisms—based on pheromone signals and direct interaction among individuals—seem to be responsible for other types of collective behavior in ants and other social insects, such as the construction of bridges or shelters formed of ants’ bodies described in chapter 1, although many aspects of such behavior are still not very well understood.
Biological Metabolism
Metabolism is the group of chemical processes by which living organisms use the energy they take in from food, air, or sunlight to maintain all the functions needed for life. These chemical processes occur largely inside of cells, via chains of chemical reactions called metabolic pathways. In every cell of an organism’s body, nutrient molecules are processed to yield energy, and cellular components are built up via parallel metabolic pathways. These components are needed for internal maintenance and repair and for external functions and intercellular communication. At any given time, millions of molecules in the cell drift around randomly in the cytoplasm. The molecules continually encounter one another. Occasionally (on a scale of microseconds), enzymes encounter molecules of matching shape, speeding up the chemical reactions the enzymes control. Sequences of such reactions cause large molecules to be built up gradually.
Just as lymphocytes affect immune system dynamics by releasing cytokines, and as ants affect foraging behavior by releasing pheromones, chemical reactions that occur along a metabolic pathway continually change the speed of and resources given to that particular pathway.
In general, metabolic pathways are complex sequences of chemical reactions, controlled by self-regulating feedback. Glycolysis is one example of a metabolic pathway that occurs in all life forms—it is a multistep process in which glucose is transformed into the chemical pryruvate, which is then used by the metabolic pathway called the citric acid cycle to produce, among other things, the molecule called ATP (adenosine triphosphate), which is the principal source of usable energy in a cell.
At any given time, hundreds of such pathways are being followed, some independent, some interdependent. The pathways result in new molecules, initiation of other metabolic pathways, and the regulation of themselves or other metabolic pathways.
Similar to the regulation mechanisms I described above for the immune system and ant colonies, metabolic regulation mechanisms are based on feedback. Glycolysis is a great example of this. One of the main purposes of glycolysis is to provide chemicals necessary for the creation of ATP. If there is a large amount of ATP in the cell, this slows down the rate of glycolysis and thus decreases the rate of new ATP production. Conversely, when the cell is lacking in ATP, the rate of glycolysis goes up. In general, the speed of a metabolic pathway is often regulated by the chemicals that are produced by that pathway.
Information Processing in These Systems
Let me now attempt to answer the questions about information processing I posed at the beginning of this chapter:
· What plays the role of “information” in these systems?
· How is it communicated and processed?
· How does this information acquire meaning? And to whom?
WHAT PLAYS THE ROLE OF INFORMATION?
As was the case for cellular automata, when I talk about information processing in these systems I am referring not to the actions of individual components such as cells, ants, or enzymes, but to the collective actions of large groups of these components. Framed in this way, information is not, as in a traditional computer, precisely or statically located in any particular place in the system. Instead, it takes the form of statistics and dynamics of patterns over the system’s components.
In the immune system the spatial distribution and temporal dynamics of lymphocytes can be interpreted as a dynamic representation of information about the continually changing population of pathogens in the body. Similarly, the spatial distribution and dynamics of cytokine concentrations encode large-scale information about the immune system’s success in killing pathogens and avoiding harm to the body.
In ant colonies, information about the colony’s food environment is represented, in a dynamic way, by the statistical distribution of ants on various trails. The colony’s overall state is represented by the dynamic distribution of ants performing different tasks.
In cellular metabolism information about the current state and needs of the cell are continually reflected in the spatial concentrations and dynamics of different kinds of molecules.
HOW IS INFORMATION COMMUNICATED AND PROCESSED?
Communication via Sampling
One consequence of encoding information as statistical and time-varying patterns of low-level components is that no individual component of the system can perceive or communicate the “big picture” of the state of the system. Instead, information must be communicated via spatial and temporal sampling.
In the immune system, for example, lymphocytes sample their environment via receptors for both antigens and signals from other immune system cells in the form of cytokines. It is the results of the lymphocytes’ samples of the spatial and temporal concentration of these molecular signals that cause lymphocytes to become active or stay dormant. Other cells are in turn affected by the samples they take of the concentration and type of active lymphocytes, which can lead pathogen-killer cells to particular areas in the body.
In ant colonies, an individual ant samples pheromone signals via its receptors. It bases its decisions on which way to move on the results of these sampled patterns of concentrations of pheromones in its environment. As I described above, individual ants also use sampling of concentration-based information—via random encounters with other ants—to decide when to adopt a particular task. In cellular metabolism, feedback in metabolic pathways arises from bindings between enzymes and particular molecules as enzymes sample spatial and time-varying concentrations of molecules.
Random Components of Behavior
Given the statistical nature of the information read, the actions of the system need to have random (or at least “unpredictable”) components. All three systems described above use randomness and probabilities in essential ways. The receptor shape of each individual lymphocyte has a randomly generated component so as to allow sampling of many possible shapes. The spatial pattern of lymphocytes in the body has a random component due to the distribution of lymphocytes by the bloodstream so as to allow sampling of many possible spatial patterns of antigens. The detailed thresholds for activation of lymphocytes, their actual division rates, and the mutations produced in the offspring all involve random aspects.
Similarly, the movements of ant foragers have random components, and these foragers encounter and are attracted to pheromone trails in a probabilistic way. Ants also task-switch in a probabilistic manner. Biochemist Edward Ziff and science historian Israel Rosenfield describe this reliance on randomness as follows: “Eventually, the ants will have established a detailed map of paths to food sources. An observer might think that the ants are using a map supplied by an intelligent designer of food distribution. However, what appears to be a carefully laid out mapping of pathways to food supplies is really just a consequence of a series of random searches.”
Cellular metabolism relies on random diffusion of molecules and on probabilistic encounters between molecules, with probabilities changing as relative concentrations change in response to activity in the system.
It appears that such intrinsic random and probabilistic elements are needed in order for a comparatively small population of simple components (ants, cells, molecules) to explore an enormously larger space of possibilities, particularly when the information to be gained from such explorations is statistical in nature and there is little a priori knowledge about what will be encountered.
However, randomness must be balanced with determinism: self-regulation in complex adaptive systems continually adjusts probabilities of where the components should move, what actions they should take, and, as a result, how deeply to explore particular pathways in these large spaces.
Fine-Grained Exploration
Many, if not all, complex systems in biology have a fine-grained architecture, in that they consist of large numbers of relatively simple elements that work together in a highly parallel fashion.
Several possible advantages are conferred by this type of architecture, including robustness, efficiency, and evolvability. One additional major advantage is that a fine-grained parallel system is able to carry out what Douglas Hofstadter has called a “parallel terraced scan.” This refers to a simultaneous exploration of many possibilities or pathways, in which the resources given to each exploration at a given time depend on the perceived success of that exploration at that time. The search is parallel in that many different possibilities are explored simultaneously, but is “terraced” in that not all possibilities are explored at the same speeds or to the same depth. Information is used as it is gained to continually reassess what is important to explore.
For example, at any given time, the immune system must determine which regions of the huge space of possible pathogen shapes should be explored by lymphocytes. Each of the trillions of lymphocytes in the body at any given time can be seen as a particular mini-exploration of a range of shapes. The shape ranges that are most successful (i.e., bind strongly to antigens) are given more exploration resources, in the form of mutated offspring lymphocytes, than the shape ranges that do not pan out (i.e., lymphocytes that do not bind strongly). However, while exploiting the information that has been obtained, the immune system continues at all times to generate new lymphocytes that explore completely novel shape ranges. Thus the system is able to focus on the most promising possibilities seen so far, while never neglecting to explore new possibilities.
Similarly, ant foraging uses a parallel-terraced-scan strategy: many ants initially explore random directions for food. If food is discovered in any of these directions, more of the system’s resources (ants) are allocated, via the feedback mechanisms described above, to explore those directions further. At all times, different paths are dynamically allocated exploration resources in proportion to their relative promise (the amount and quality of the food that has been discovered at those locations). However, due to the large number of ants and their intrinsic random elements, unpromising paths continue to be explored as well, though with many fewer resources. After all, who knows—a better source of food might be discovered.
In cellular metabolism such fine-grained explorations are carried out by metabolic pathways, each focused on carrying out a particular task. A pathway can be speeded up or slowed down via feedback from its own results or from other pathways. The feedback itself is in the form of time-varying concentrations of molecules, so the relative speeds of different pathways can continually adapt to the moment-to-moment needs of the cell.
Note that the fine-grained nature of the system not only allows many different paths to be explored, but it also allows the system to continually change its exploration paths, since only relatively simple micro-actions are taken at any time. Employing more coarse-grained actions would involve committing time to a particular exploration that might turn out not to be warranted. In this way, the fine-grained nature of exploration allows the system to fluidly and continuously adapt its exploration as a result of the information it obtains. Moreover, the redundancy inherent in fine-grained systems allows the system to work well even when the individual components are not perfectly reliable and the information available is only statistical in nature. Redundancy allows many independent samples of information to be made, and allows fine-grained actions to be consequential only when taken by large numbers of components.
Interplay of Unfocused and Focused Processes
In all three example systems there is a continual interplay of unfocused, random explorations and focused actions driven by the system’s perceived needs.
In the immune system, unfocused explorations are carried out by a continually changing population of lymphocytes with different receptors, collectively prepared to approximately match any antigen. Focused explorations consist of the creation of offspring that are variations of successful lymphocytes, which allow these explorations to zero in on a particular antigen shape.
Likewise, ant foraging consists of unfocused explorations by ants moving at random, looking for food in any direction, and focused explorations in which ants follow existing pheromone trails.
In cellular metabolism, unfocused processes of random exploration by molecules are combined with focused activation or inhibition driven by chemical concentrations and genetic regulation.
As in all adaptive systems, maintaining a correct balance between these two modes of exploring is essential. Indeed, the optimal balance shifts over time. Early explorations, based on little or no information, are largely random and unfocused. As information is obtained and acted on, exploration gradually becomes more deterministic and focused in response to what has been perceived by the system. In short, the system both explores to obtain information and exploits that information to successfully adapt. This balancing act between unfocused exploration and focused exploitation has been hypothesized to be a general property of adaptive and intelligent systems. John Holland, for example, has cited this balancing act as a way to explain how genetic algorithms work.
HOW DOES INFORMATION ACQUIRE MEANING?
How information takes on meaning (some might call it purpose) is one of those slippery topics that has filled many a philosophy tome over the eons. I don’t think I can add much to what the philosophers have said, but I do claim that in order to understand information processing in living systems we will need to answer this question in some form.
In my view, meaning is intimately tied up with survival and natural selection. Events that happen to an organism mean something to that organism if those events affect its well-being or reproductive abilities. In short, the meaning of an event is what tells one how to respond to it. Similarly, events that happen to or within an organism’s immune system have meaning in terms of their effects on the fitness of the organism. (I’m using the term fitness informally here.) These events mean something to the immune system because they tell it how to respond so as to increase the organism’s fitness—similarly with ant colonies, cells, and other information-processing systems in living creatures. This focus on fitness is one way I can make sense of the notion of meaning and apply it to biological information-processing systems.
But in a complex system such as those I’ve described above, in which simple components act without a central controller or leader, who or what actually perceives the meaning of situations so as to take appropriate actions? This is essentially the question of what constitutes consciousness or self-awareness in living systems. To me this is among the most profound mysteries in complex systems and in science in general. Although this mystery has been the subject of many books of science and philosophy, it has not yet been completely explained to anyone’s satisfaction.
Thinking about living systems as doing computation has had an interesting side effect: it has inspired computer scientists to write programs that mimic such systems in order to accomplish real-world tasks. For example, ideas about information processing in the immune system has inspired so-called artificial immune systems: programs that adaptively protect computers from viruses and other intruders. Similarly ant colonies have inspired what are now called “ant colony optimization algorithms,” which use simulated ants, secreting simulated pheromones and switching between simulated jobs, to solve hard problems such as optimal cell-phone communications routing and optimal scheduling of delivery trucks. I don’t know of any artificial intelligence programs inspired both by these two systems and by cellular metabolism, except for one I myself wrote with my Ph.D. advisor, which I describe in the next chapter.